Description
Usage
Arguments
Example Output
See Also
Examples
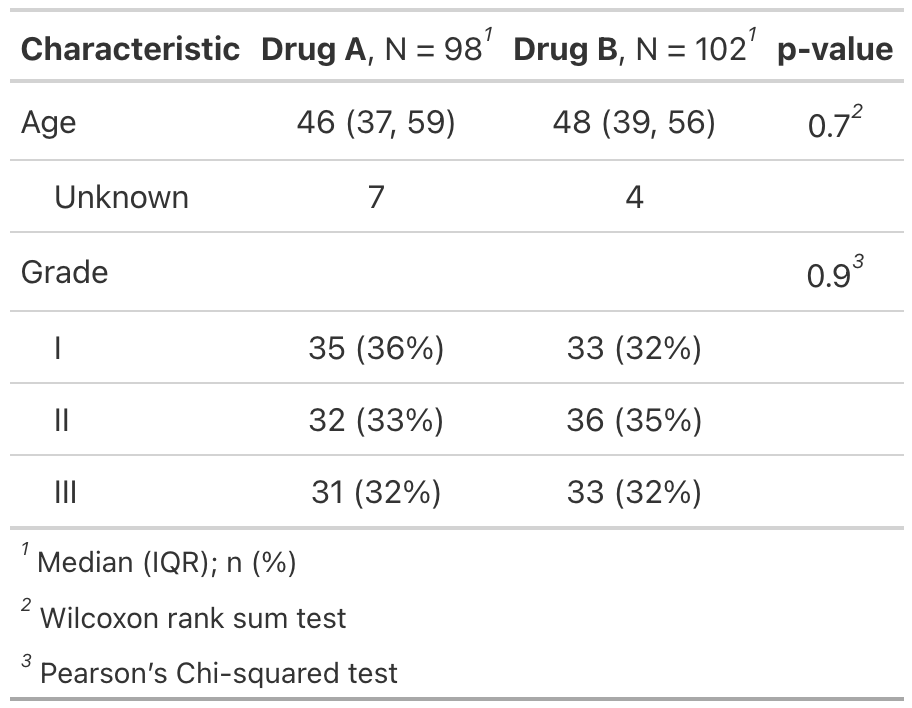
Run this code# Example 1 ----------------------------------
separate_p_footnotes_ex1 <-
trial %>%
select(trt, age, grade) %>%
tbl_summary(by = trt) %>%
add_p() %>%
separate_p_footnotes()
Run the code above in your browser using DataLab